Research
You can also find my articles on my Google Scholar profile.
Nanopore signal classification with pangenome indexes
Nanopore sequencing generates reads by measuring electrical current signal that is converted to nucleic acid sequences typically with a neural network. This basecalling step is a bottleneck in real-time classification pipelines. We developed a novel nanopore signal-based read classification method that uses the r-index, a full-text index that scales to pangenomes. This method, Sigmoni, is significantly faster and more accurate than existing methods for classifying nanopore reads against large pangenomes.
Sigmoni paper (pdf), published in 2024 in Bioinformatics (ISMB 2024)
🏆 RECOMB-seq Best Poster/Short Talk - RECOMB-seq talk recording, in Istanbul, Türkiye (April 2023)
🏆 ISMB HitSeq Best Talk - ISMB talk recording and slides, in Montreal, Quebec (July 2024)
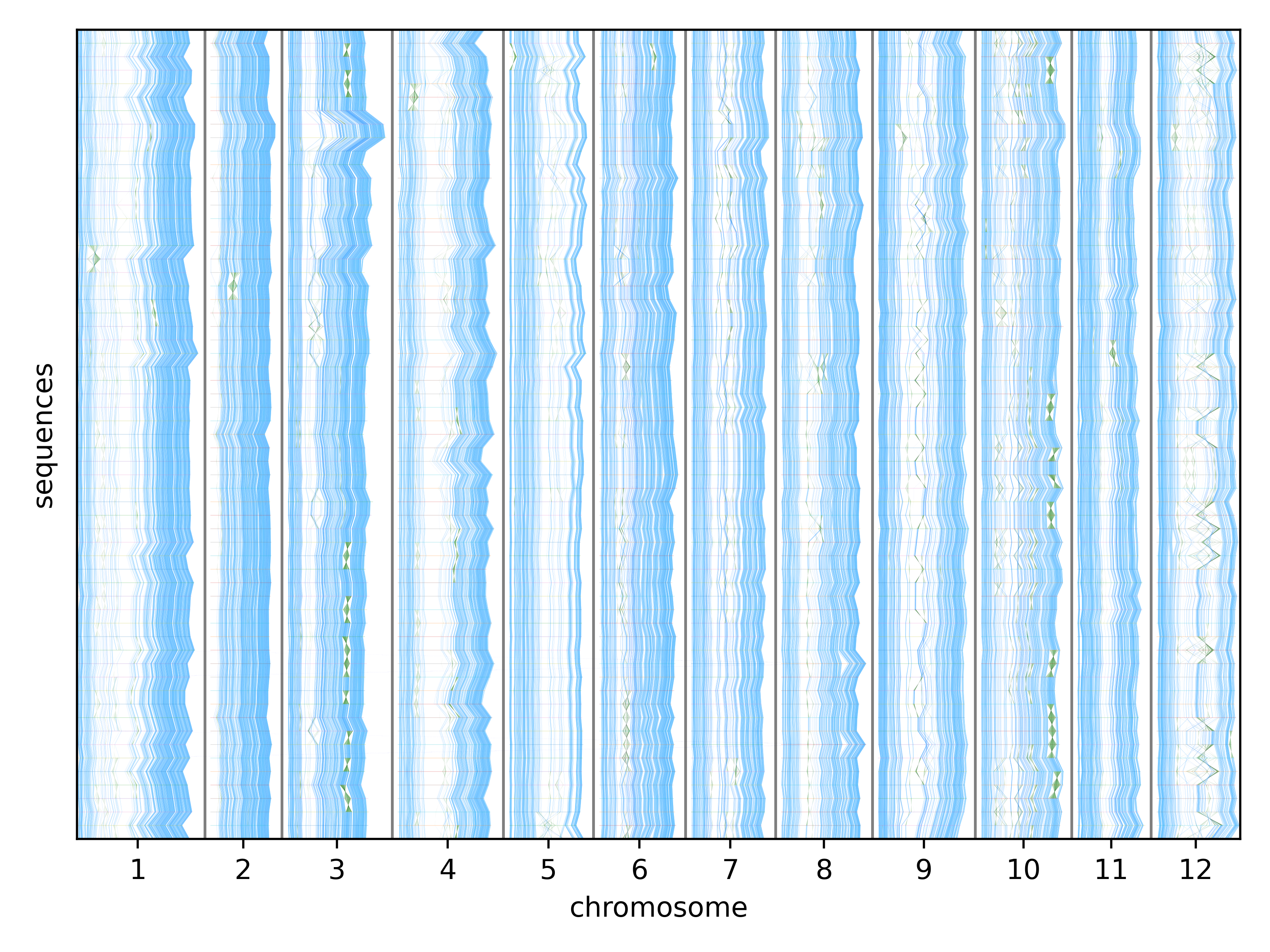
Building and visualizing pangenomes
 Pangenomes require an underlying alignment for interpretability and usability. This structure can take the form of a multiple alignment or graph, but these are impractical to compute. We developed a novel method, Mumemto, to compute maximal unique matches (multi-MUMs), commonly used as anchors for alignment, at the scale of hundreds of human genomes. Mumemto can visualize pangenome synteny (example below), accelerate graph construction, identify misassemblies, and even improve full-text index-based classification (see further work).
Pangenomes require an underlying alignment for interpretability and usability. This structure can take the form of a multiple alignment or graph, but these are impractical to compute. We developed a novel method, Mumemto, to compute maximal unique matches (multi-MUMs), commonly used as anchors for alignment, at the scale of hundreds of human genomes. Mumemto can visualize pangenome synteny (example below), accelerate graph construction, identify misassemblies, and even improve full-text index-based classification (see further work).
Mumemto paper (open access), published in 2025 in Genome Biology
🏆 ISMB EvolCompGen Best Poster - poster (large file, 25 MB), in Liverpool, UK (July 2025)
Follow-up paper available on BioRxiv : Partitioned Multi-MUM finding for scalable pangenomics
Software on github